Proteins perform without water
14.02.2013
The thin film of water around proteins, their hydration water, is vital to the macromolecule’s biological activity. It was believed that without hydration water, proteins would not only be incorrectly folded but also lack the conformational flexibility that animates their 3D structures and brings them to life. However, a team of scientists from the Institut de Biologie Structurale (IBS), the University of Bristol, the Australian National University, the Jülich Centre for Neutron Scattering (JCNS) at FRM II and the Institut Laue-Langevin (ILL) found out otherwise.
They investigated novel nano-materials that are hybrids of proteins surrounded by a ‘corona’ of polymer surfactant molecules (see figure) that contain no water, nor any other solvent for that matter. Yet, surprisingly, these nano-hybrids are biologically active. Furthermore, researchers from the IBS, the University of California, the Australian Institute of Science and Technology Organisation and the JCNS have applied neutron scattering techniques to an intrinsically unfolded protein called tau and found out that protein and hydration water motions mutually affect and shape each other.
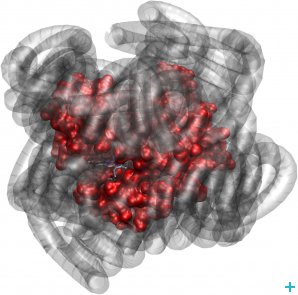
Myoglobin (red) exhibits biologically relevant dynamics, even when its hydration sphere is replaced by a polymer surfactant corona (grey)
Myoglobin behaviour without water
Myoglobin is common to almost all mammals and responsible for the red colour of raw meat. Observing biological activity in solvent-free protein polymer hybrids means that existing theories about aqueous and non-aqueous solvents being unique promoters of protein dynamics linked to function might have to be reconsidered.
As a first step of their research, these scientists wanted to assess whether the structure of myoglobin could still move and continue to bind oxygen if all the water was completely removed and replaced by synthetic molecules. The team analysed three samples: a wet sample (the protein in water), a dry sample (the dehydrated protein) and a dry protein-polymer hybrid sample.
What they found was that the myoglobin molecules surrounded by polymer moved just as well as the wet sample, and that the dry sample had very little mobility. These observations lead to the conclusion that the polymer surfactant coating plasticises protein structures in a similar way to hydration water.
Knowing that proteins can function outside of water opens them up to use in real life applications because it shows that there are other alternatives if water is unavailable. Examples of where they could be used include biochemical gas sensors, as myoglobin can bind carbon monoxide molecules.
Another potential application is in the development of new wound dressings, where the liquid protein could be applied to the wound to reduce healing time by supplying oxygen or other essential chemicals to the damaged tissue.
Adam Perriman of the University of Bristol said: “These discoveries have increased our fundamental understanding of proteins and how they behave, which could create many new opportunities for their application in industrial processing and in medical technologies. The fact that our proteins can happily perform their function outside of water, a substance generally thought to be vital for life, really drives home just how robust these biological nanomachines are.”
Martin Weik of the IBS explained: “Neutron scattering techniques are excellent for studying the dynamics of proteins and of their environment. The world class neutron scattering facilities at the ILL and the FRM II allow us to analyse how proteins move, thus complementing the single snapshots of their structures provided by crystallography.”
Intrinsically disordered proteins – the tau protein
These researchers have extended their studies to a third class of proteins, i.e. intrinsically disordered proteins (IDP) that do not have a well defined 3D structure and fold only upon interaction with a target ligand. Most proteins can only fulfill their intended function when the chains of amino acids that constitute them are folded into a convoluted cluster in the right way for the individual protein. However, the tau protein, which stabilizes transport paths in biological cells, is only partially folded. Disordered tau proteins tend to build up as deposits and destroy cells. Such deposits are found in the brains of those suffering from dementia, for example, Alzheimer patients.
Very little is known about the functionality of disordered proteins. Now, for the first time, the team has investigated the motions of the tau protein and its hydration shell as representative of disordered proteins. They wanted to understand how its flexibility and its interactions with water differ from ordered proteins from cell plasma and the cell membrane. Combining neutron scattering and protein perdeuteration, they found that the coupling of the disordered tau protein with water motions was much tighter than for folded proteins. They also revealed a greater motional flexibility and more restricted water motions on the IDP surface, as compared to folded proteins. The results provide evidence that protein and hydration-water motions mutually affect and shape each other, and that there is a gradient of coupling across different protein classes that may play a functional role in macromolecular activity in a cellular context.
IDPs are of significant interest in a medical context because they can aggregate and cluster together to create the amyloid fibrils behind neuro-degenerative diseases such as Parkinson’s and Alzheimer’s. Whilst the ordered structure of folded proteins makes it possible to develop drugs that fit into the protein like a key in a lock, the conformational variability of an intrinsically disordered protein like tau makes it more difficult. A more in-depth understanding of their dynamics is required and the discovery of tight coupling with water motions is a significant step forward.
This article was prepared from materials from the JCNS, ILL and PSB. NMI3 thanks Martin Weik from the Institut de Biologie Structurale for reviewing it.
Original publications
- Gallat F.-X., Laganowski A., Wood K., Gabel F., van Eijck L., Wuttke J., Moulin M., Härtlein M., Eisenberg D., Colletier J.-P., Zaccai G., Weik M. (2012) Dynamical Coupling of Intrinsically Disordered Proteins and Their Hydration Water: Comparison with Folded Soluble and Membrane Proteins. Biophys J., 103(1), 129–136
- Gallat F. X., Brogan A. P., Fichou Y., McGrath N., Moulin M., Hartlein M., Combet J., Wuttke J., Mann S., Zaccai G., Jackson C. J., Perriman A. W., Weik M. (2012) A Polymer Surfactant Corona Dynamically Replaces Water in Solvent-Free Protein Liquids and Ensures Macromolecular Flexibility and Activity. J. Am. Chem. Soc., 134, 13168-13171
For a more complete description, please read Issue 4 of our newsletter.
